Protein and protein complexes are fascinating nanomachines performing key functions in living organisms. Improved over billion years of natural selection, protein complexes are fine-tuned by evolution to perform their function with unmatchable efficiency, and complex physicochemical mechanism are exploited by nature to control and modify their activity. Single-molecule fluorescence techniques allow us to directly look at these molecules while performing their activity, and therefore, to extract information about the conformational changes occurring during their inner working, or in response to external factors.
Cellulose is the most abundant organic molecule on earth, and it is a huge natural reservoir of energy and carbon that can be potentially used for the production of biofuels and chemicals. Despite their availability, the chemical and structural heterogeneous nature of lignocellulosic materials hamper their enzymatic degradation. Cellulosomes are huge extracellular protein complexes produced by some bacteria with unmatched efficiency in the degradation of cellulosic materials. Using single-molecule fluorescence resonance energy transfer (smFRET), we study the assembly, dynamics and structural regulation of this captivating nanomachines.


Figure 1. a) The Cellulosome of Clostridium thermocellum. b) Detail of the two modes of binding of the cellulosome cohesin-dockerin interaction.
Furthermore, we have used smFRET to study the regulation and conformational changes of the transcription machinery of archaea and eukaryotic cells. Forces are ubiquitous during the work cycle of RNA polymerases and accessories proteins, and therefore any description of these systems that does not take them into account is largely incomplete. Accordingly, we have developed a totally new type of force spectroscopy method that uses a DNA origami-based force clamp to exert piconewton forces in these systems. Unlike classical force spectroscopy methods, as AFM and optical tweezers, our force probe is of nanometric scale and a fluorescence-base readout allows for measurement of hundreds of molecules simultaneously.
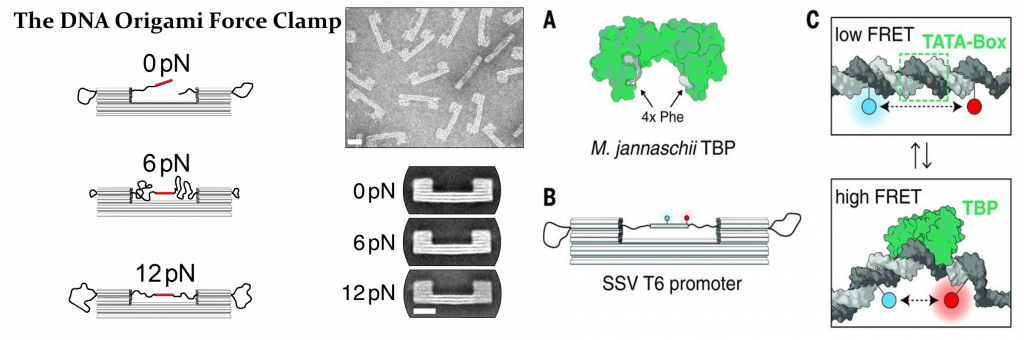
Figure 2. DNA-origami based force actuator to study the DNA transcription machinery.
Besides, we have recently developed a totally new branch of light-activatable enzymes for biotechnological use, including mesophilic and thermophilic DNA polymerases, and reverse-transcriptases. Unlike previous technologies, our new approach allows for the design of new enzymes within weeks, with applications in isothermal amplification, hot start PCR, and RT-qPCR methods. Moreover, we are currently developing DNA origami nanoantenna-based bioassays to visualize microsecond dynamics of biomolecules at high photon count rates, new MINFLUX-based super resolution microscopy techniques to study cellular processes, and we aim to exploit an exotic behavior of graphene for biophysical applications.
Z. Liu et al., High Force Catch Bond Mechanism of Bacterial Adhesion in the Human Gut. 2020.2001.2021.913590 (2020).
A. M. Vera et al., smFRET Detects Dual Binding Modes Modulated by Proline Isomerization in a Mega-Dalton Multi-Enzyme Complex. 2019.2012.2019.882373 (2019).
S. Schulz et al., TFE and Spt4/5 open and close the RNA polymerase clamp during the transcription cycle. 113, E1816-E1825 (2016).
C. Sheppard et al., Repression of RNA polymerase by the archaeo-viral regulator ORF145/RIP. Nature Communications 7, 13595 (2016).
K. Kramm et al., DNA origami-based single-molecule force spectroscopy elucidates RNA Polymerase III pre-initiation complex stability. Nature Communications 11, 2828 (2020).
P. C. Nickels et al., Molecular force spectroscopy with a DNA origami-based nanoscopic force clamp. Science (New York, N.Y.) 354, 305-307 (2016).
I. Kaminska et al., Distance Dependence of Single-Molecule Energy Transfer to Graphene Measured with DNA Origami Nanopositioners. Nano Letters 19, 4257-4262 (2019).