









Group theses
2025
Philip Tinnefeld; Samrat Basak
Polymer dots for nanoscale live-cell imaging Journal Article
In: Nature Photonics, 2025.
@article{nokey,
title = {Polymer dots for nanoscale live-cell imaging},
author = {Philip Tinnefeld and Samrat Basak},
url = {https://doi.org/10.1038/s41566-025-01811-0
},
doi = {10.1038/s41566-025-01811-0},
year = {2025},
date = {2025-12-02},
urldate = {2025-12-02},
journal = {Nature Photonics},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Martina Pfeiffer; Fiona Cole; Dongfang Wang; Yonggang Ke; Philip Tinnefeld
Spring-loaded DNA origami arrays as energy-supplied hardware for modular nanorobots Journal Article
In: Science Robotics, 2025.
@article{nokey,
title = {Spring-loaded DNA origami arrays as energy-supplied hardware for modular nanorobots},
author = {Martina Pfeiffer and Fiona Cole and Dongfang Wang and Yonggang Ke and Philip Tinnefeld},
url = {https://www.science.org/doi/10.1126/scirobotics.adu3679},
doi = {10.1126/scirobotics.adu3679},
year = {2025},
date = {2025-10-22},
journal = {Science Robotics},
keywords = {},
pubstate = {published},
tppubtype = {article}
}

Renukka Yaadav; Kateryna Trofymchuk; Mihir Dass; Vivien Behrendt; Benedikt Hauer; Jan Schütz; Cindy Close; Michael Scheckenbach; Giovanni Ferrari; Leoni Mäurer; Sophia Sebina; Viktorija Glembockyte; Tim Liedl; Philip Tinnefeld
Bringing Attomolar Detection to the Point-of-Care with Nanopatterned DNA Origami Nanoantennas Journal Article
In: Advanced Materials, 2025.
@article{nokey,
title = {Bringing Attomolar Detection to the Point-of-Care with Nanopatterned DNA Origami Nanoantennas},
author = {Renukka Yaadav and Kateryna Trofymchuk and Mihir Dass and Vivien Behrendt and Benedikt Hauer and Jan Schütz and Cindy Close and Michael Scheckenbach and Giovanni Ferrari and Leoni Mäurer and Sophia Sebina and Viktorija Glembockyte and Tim Liedl and Philip Tinnefeld},
url = { https://doi.org/10.1002/adma.202507407},
doi = {10.1002/adma.202507407},
year = {2025},
date = {2025-07-26},
journal = {Advanced Materials},
keywords = {},
pubstate = {published},
tppubtype = {article}
}

Michael Scheckenbach; Gereon Andreas Brüggenthies; Tim Schröder; Karina Betuker; Lea Wassermann; Philip Tinnefeld; Amelie Heuer-Jungemann; Viktorija Glembockyte
Monitoring the Coating of Single DNA Origami Nanostructures with a Molecular Fluorescence Lifetime Sensor Journal Article
In: Small, 2025.
@article{nokey,
title = {Monitoring the Coating of Single DNA Origami Nanostructures with a Molecular Fluorescence Lifetime Sensor},
author = {Michael Scheckenbach and Gereon Andreas Brüggenthies and Tim Schröder and Karina Betuker and Lea Wassermann and Philip Tinnefeld and Amelie Heuer-Jungemann and Viktorija Glembockyte},
url = { https://doi.org/10.1002/smll.202501044},
doi = {10.1002/smll.202501044},
year = {2025},
date = {2025-06-19},
journal = {Small},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Claudia Bastl; Cindy Close; Ingo Holtz; Blaise Gatin-Fraudet; Mareike Eis; Michelle Werum; Smilla Konrad; Laura Kneller; Killian Roßmann; Christiane Huhn; Souvik Ghosh; Julia Ast; Dorien A. Roosen; Martin Lehmann; Volker Haucke; Luc Reymond; David J. Hodson; Philip Tinnefeld; Kai Johnsson; Viktorija Glembockyte; Nicole Kilian; Johannes Broichhagen
A Silicon Rhodamine-fused Glibenclamide to Label and Detect Malaria-infected Red Blood Cells Journal Article
In: ChemBioChem, 2025, ISSN: 1439-4227.
@article{nokey,
title = {A Silicon Rhodamine-fused Glibenclamide to Label and Detect Malaria-infected Red Blood Cells},
author = {Claudia Bastl and Cindy Close and Ingo Holtz and Blaise Gatin-Fraudet and Mareike Eis and Michelle Werum and Smilla Konrad and Laura Kneller and Killian Roßmann and Christiane Huhn and Souvik Ghosh and Julia Ast and Dorien A. Roosen and Martin Lehmann and Volker Haucke and Luc Reymond and David J. Hodson and Philip Tinnefeld and Kai Johnsson and Viktorija Glembockyte and Nicole Kilian and Johannes Broichhagen},
doi = {https://doi.org/10.1002/cbic.202400628},
issn = {1439-4227},
year = {2025},
date = {2025-02-25},
urldate = {2025-02-25},
journal = {ChemBioChem},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Michael Scheckenbach; Cindy Close; Julian Bauer; Lennart Grabenhorst; Fiona Cole; Jens Köhler; Siddharth S. Matikonda; Lei Zhang; Thorben Cordes; Martin J. Schnermann; Andreas Herrmann; Philip Tinnefeld; Alan M. Szalai; Viktorija Glembockyte
Minimally Invasive DNA-Mediated Photostabilization for Extended Single-Molecule and Super-resolution Imaging Journal Article
In: bioRxiv preprint, 2025.
@article{nokey,
title = {Minimally Invasive DNA-Mediated Photostabilization for Extended Single-Molecule and Super-resolution Imaging},
author = {Michael Scheckenbach and Cindy Close and Julian Bauer and Lennart Grabenhorst and Fiona Cole and Jens Köhler and Siddharth S. Matikonda and Lei Zhang and Thorben Cordes and Martin J. Schnermann and Andreas Herrmann and Philip Tinnefeld and Alan M. Szalai and Viktorija Glembockyte},
doi = {https://doi.org/10.1101/2025.01.08.631860},
year = {2025},
date = {2025-01-10},
journal = {bioRxiv preprint},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
2024
Deepak Deepak; Jiaojiao Wu; Valentina Corvaglia und Lars Allmendinger; Michael Scheckenbach; Philip Tinnefeld; Ivan Huc
DNA Mimic Foldamer Recognition of a Chromosomal Protein Journal Article
In: Angewandte Chemie International Edition, vol. 64, iss. 8, 2024.
@article{nokey,
title = {DNA Mimic Foldamer Recognition of a Chromosomal Protein},
author = {Deepak Deepak and Jiaojiao Wu and Valentina Corvaglia und Lars Allmendinger and Michael Scheckenbach and Philip Tinnefeld and Ivan Huc},
doi = { https://doi.org/10.1002/anie.202422958},
year = {2024},
date = {2024-12-23},
journal = {Angewandte Chemie International Edition},
volume = {64},
issue = {8},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Na-Na Sun; Philip Tinnefeld; Guoliang Li; Zhike He; Qinfeng Xu
Aptamer melting biosensors for thousands of signaling and regenerating cycles Journal Article
In: Biosensors and Bioelectronics, vol. 271, pp. 116998, 2024, ISSN: 0956-5663.
@article{Tinnefeld_2024,
title = {Aptamer melting biosensors for thousands of signaling and regenerating cycles},
author = {Na-Na Sun and Philip Tinnefeld and Guoliang Li and Zhike He and Qinfeng Xu},
doi = {https://doi.org/10.1016/j.bios.2024.116998},
issn = {0956-5663},
year = {2024},
date = {2024-11-26},
journal = {Biosensors and Bioelectronics},
volume = {271},
pages = {116998},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Alan M. Szalai; Giovanni Ferrari; Lars Richter; Jakob Hartmann; Merve-Zeynep Kesici; Bosong Ji; Kush Coshic; Martin R. J. Dagleish; Annika Jaeger; Aleksei Aksimentiev; Ingrid Tessmer; Izabela Kamińska; Andrés M. Vera; Philip Tinnefeld
Single-molecule dynamic structuralbiology with vertically arranged DNA on afluorescence microscope Journal Article
In: Nature Methods, 2024.
@article{Szalai2024,
title = {Single-molecule dynamic structuralbiology with vertically arranged DNA on afluorescence microscope},
author = {Alan M. Szalai and Giovanni Ferrari and Lars Richter and Jakob Hartmann and Merve-Zeynep Kesici and Bosong Ji and Kush Coshic and Martin R. J. Dagleish and Annika Jaeger and Aleksei Aksimentiev and Ingrid Tessmer and Izabela Kamińska and Andrés M. Vera and Philip Tinnefeld},
url = {https://rdcu.be/dZyyO },
doi = {https://doi.org/10.1038/s41592-024-02498-x},
year = {2024},
date = {2024-11-08},
urldate = {2024-11-08},
journal = {Nature Methods},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Lennart Grabenhorst; Martina Pfeiffer; Thea Schinkel; Mirjam Kümmerlin; Gereon A. Brüggenthies; Jasmin B. Maglic; Florian Selbach; Alexander T. Murr; Philip Tinnefeld; Viktorija Glembockyte
Engineering modular and tunablesingle-molecule sensors by decouplingsensing from signal output Journal Article
In: Nature Nanotechnology, 2024, ISSN: 1748-3395.
@article{Grabenhorst2024,
title = {Engineering modular and tunablesingle-molecule sensors by decouplingsensing from signal output},
author = {Lennart Grabenhorst and Martina Pfeiffer and Thea Schinkel and Mirjam Kümmerlin and Gereon A. Brüggenthies and Jasmin B. Maglic and Florian Selbach and Alexander T. Murr and Philip Tinnefeld and Viktorija Glembockyte },
doi = {https://doi.org/10.1038/s41565-024-01804-0},
issn = {1748-3395},
year = {2024},
date = {2024-11-07},
journal = {Nature Nanotechnology},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Michael Scheckenbach; Gereon Andreas Brüggenthies; Tim Schröder; Karina Betuker; Lea Wassermann; Philip Tinnefeld; Amelie Heuer-Jungemann; Viktorija Glembockyte
Monitoring the Coating of Single DNA Origami Nanostructures with a Molecular Fluorescence Lifetime Sensor Journal Article
In: bioRxiv preprint, 2024.
@article{nokey,
title = {Monitoring the Coating of Single DNA Origami Nanostructures with a Molecular Fluorescence Lifetime Sensor},
author = {Michael Scheckenbach and Gereon Andreas Brüggenthies and Tim Schröder and Karina Betuker and Lea Wassermann and Philip Tinnefeld and Amelie Heuer-Jungemann and Viktorija Glembockyte},
url = {https://doi.org/10.1101/2024.10.28.620667},
doi = {10.1101/2024.10.28.620667},
year = {2024},
date = {2024-10-31},
urldate = {2024-10-31},
journal = {bioRxiv preprint},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Lorena Manzanares; Dahnan Spurling; Alan M. Szalai; Tim Schröder; Ece Büber; Giovanni Ferrari; Martin R. J. Dagleish; Valeria Nicolosi; Philip Tinnefeld
In: Advanced Materials, 2024.
@article{nokey,
title = {2D Titanium Carbide MXene and Single-Molecule Fluorescence: Distance-Dependent Nonradiative Energy Transfer and Leaflet-Resolved Dye Sensing in Lipid Bilayers},
author = {Lorena Manzanares and Dahnan Spurling and Alan M. Szalai and Tim Schröder and Ece Büber and Giovanni Ferrari and Martin R. J. Dagleish and Valeria Nicolosi and Philip Tinnefeld},
url = {https://doi.org/10.1002/adma.202411724},
doi = {10.1002/adma.202411724},
year = {2024},
date = {2024-10-24},
urldate = {2024-10-24},
journal = {Advanced Materials},
keywords = {},
pubstate = {published},
tppubtype = {article}
}

Renukka Yaadav; Kateryna Trofymchuk; Mihir Dass; Vivien Behrendt; Benedikt Hauer; Jan Schütz; Cindy Close; Michael Scheckenbach; Giovanni Ferrari; Leoni Maeurer; Sophia Sebina; Viktorija Glembockyte; Tim Liedl; Philip Tinnefeld
Bringing Attomolar Detection to the Point-of-Care with Nanopatterned DNA Origami Nanoantennas Journal Article
In: bioRxiv preprint, 2024.
@article{nokey,
title = {Bringing Attomolar Detection to the Point-of-Care with Nanopatterned DNA Origami Nanoantennas},
author = {Renukka Yaadav and Kateryna Trofymchuk and Mihir Dass and Vivien Behrendt and Benedikt Hauer and Jan Schütz and Cindy Close and Michael Scheckenbach and Giovanni Ferrari and Leoni Maeurer and Sophia Sebina and Viktorija Glembockyte and Tim Liedl and Philip Tinnefeld},
url = {https://doi.org/10.1101/2024.10.14.618183},
doi = {10.1101/2024.10.14.618183},
year = {2024},
date = {2024-10-17},
urldate = {2024-10-17},
journal = {bioRxiv preprint},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Fiona Cole; Martina Pfeiffer; Dongfang Wang; Tim Schröder; Yonggang Ke; Philip Tinnefeld
Controlled mechanochemical coupling of anti-junctions in DNA origami arrays Journal Article
In: Nature Communications, 2024.
@article{nokey,
title = {Controlled mechanochemical coupling of anti-junctions in DNA origami arrays},
author = {Fiona Cole and Martina Pfeiffer and Dongfang Wang and Tim Schröder and Yonggang Ke and Philip Tinnefeld},
url = {https://www.nature.com/articles/s41467-024-51721-y#citeas},
doi = {10.1038/s41467-024-51721-y},
year = {2024},
date = {2024-09-10},
urldate = {2024-09-10},
journal = {Nature Communications},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Ece Büber; Renukka Yaadav; Tim Schröder; Henri G. Franquelim; Philip Tinnefeld
DNA Origami Vesicle Sensors with Triggered Single-Molecule Cargo Transfer Journal Article
In: Angewandte Chemie International Edition, 2024.
@article{nokey,
title = {DNA Origami Vesicle Sensors with Triggered Single-Molecule Cargo Transfer},
author = {Ece Büber and Renukka Yaadav and Tim Schröder and Henri G. Franquelim and Philip Tinnefeld},
url = {https://onlinelibrary.wiley.com/doi/10.1002/anie.202408295},
doi = {10.1002/anie.202408295},
year = {2024},
date = {2024-09-09},
urldate = {2024-09-09},
journal = {Angewandte Chemie International Edition},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Melanie Meincke; Andre Bazzone; Stephan Holzhauser; Maria Barthmes; Lars Richter; Fabian Knechtel; Evelyn Ploetz; Michael George; Niels Fertig; Izabela Kamińska; Philip Tinnefeld
Integration of highly sensitive large-area graphene-based biosensors in an automated sensing platform Journal Article
In: Measurement, vol. 240, pp. 115592, 2024.
@article{,
title = {Integration of highly sensitive large-area graphene-based biosensors in an automated sensing platform},
author = {Melanie Meincke and Andre Bazzone and Stephan Holzhauser and Maria Barthmes and Lars Richter and Fabian Knechtel and Evelyn Ploetz and Michael George and Niels Fertig and Izabela Kamińska and Philip Tinnefeld},
doi = {https://doi.org/10.1016/j.measurement.2024.115592},
year = {2024},
date = {2024-08-28},
urldate = {2024-08-28},
journal = {Measurement},
volume = {240},
pages = {115592},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Karolina Gronkiewicz; Lars Richter; Fabian Knechtel; Patryk Pyrcz; Paul Leidinger; Sebastian Günther; Evelyn Ploetz; Philip Tinnefeld; Izabela Kamińska
Expanding the range of graphene energy transfer with multilayer graphene Journal Article
In: Nanoscale, 2024.
@article{nokey,
title = {Expanding the range of graphene energy transfer with multilayer graphene},
author = {Karolina Gronkiewicz and Lars Richter and Fabian Knechtel and Patryk Pyrcz and Paul Leidinger and Sebastian Günther and Evelyn Ploetz and Philip Tinnefeld and Izabela Kamińska},
url = {https://doi.org/10.1039/D4NR01723D},
doi = {10.1039/D4NR01723D},
year = {2024},
date = {2024-06-26},
journal = {Nanoscale},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
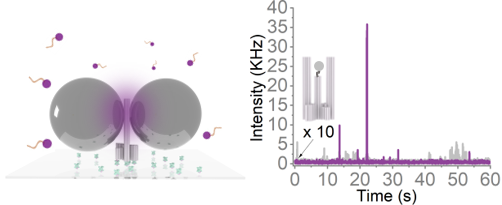
Renukka Yaadav; Kateryna Trofymchuk; Feng Gong; Xinghu Ji; Florian Steiner; Philip Tinnefeld; Zhike He
Broad-Band Fluorescence Enhancement of QDs Captured in the Hotspot of DNA Origami Nanonantennas Journal Article
In: The Journal of Physical Chemistry C, 2024.
@article{nokey,
title = {Broad-Band Fluorescence Enhancement of QDs Captured in the Hotspot of DNA Origami Nanonantennas},
author = {Renukka Yaadav and Kateryna Trofymchuk and Feng Gong and Xinghu Ji and Florian Steiner and Philip Tinnefeld and Zhike He},
doi = {doi.org/10.1021/acs.jpcc.4c01797},
year = {2024},
date = {2024-05-24},
urldate = {2024-05-24},
journal = {The Journal of Physical Chemistry C},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Zhaowei Liu, Haipei Liu; Andrés M. Vera; Byeongseon Yang; Philip Tinnefeld; Michael A. Nash
Engineering an artificial catch bond using mechanical anisotropy Journal Article
In: Nature Communications, 2024.
@article{nokey,
title = {Engineering an artificial catch bond using mechanical anisotropy},
author = {Zhaowei Liu, Haipei Liu and Andrés M. Vera and Byeongseon Yang and Philip Tinnefeld and Michael A. Nash},
url = {https://doi.org/10.1038/s41467-024-46858-9},
doi = {10.1038/s41467-024-46858-9},
year = {2024},
date = {2024-04-08},
urldate = {2024-04-08},
journal = {Nature Communications},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Julian Bauer; Fiona Cole; Renukka Yaadav; Jonas Zähringer; Tim Schröder; Philip Tinnefeld
Ultra-specific detection of nucleic acids by intramolecular referencing Journal Article
In: Single Molecule Spectroscopy and Superresolution Imaging XVII, vol. 12849, 2024.
@article{nokey,
title = {Ultra-specific detection of nucleic acids by intramolecular referencing},
author = {Julian Bauer and Fiona Cole and Renukka Yaadav and Jonas Zähringer and Tim Schröder and Philip Tinnefeld},
url = {https://doi.org/10.1117/12.3010119},
doi = {10.1117/12.3010119},
year = {2024},
date = {2024-03-12},
urldate = {2024-03-12},
journal = {Single Molecule Spectroscopy and Superresolution Imaging XVII},
volume = {12849},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Rosa Antón; Miguel Á. Treviño; David Pantoja-Uceda; Sara Félix; María Babu; Eurico J. Cabrita; Markus Zweckstetter; Philip Tinnefeld; Andrés M. Vera; Javier Oroz
Alternative low-populated conformations prompt phase transitions in polyalanine repeat expansions Journal Article
In: Nature Communications, 2024.
@article{nokey,
title = {Alternative low-populated conformations prompt phase transitions in polyalanine repeat expansions},
author = {Rosa Antón and Miguel Á. Treviño and David Pantoja-Uceda and Sara Félix and María Babu and Eurico J. Cabrita and Markus Zweckstetter and Philip Tinnefeld and Andrés M. Vera and Javier Oroz},
url = {https://doi.org/10.1038/s41467-024-46236-5},
doi = {10.1038/s41467-024-46236-5},
year = {2024},
date = {2024-03-02},
urldate = {2024-03-02},
journal = {Nature Communications},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Jonas Binder; Joshua Winkeljann; Katharina Steinegger; Lara Trnovec; Daria Orekhova; Jonas Zähringer; Andreas Hörner; Valentin Fell; Philip Tinnefeld; Benjamin Winkeljann; Wolfgang Frieß; Olivia M Merkel
Closing the Gap between Experiment and Simulation─A Holistic Study on the Complexation of Small Interfering RNAs with Polyethylenimine Journal Article
In: Molecular Pharmaceutics, 2024.
@article{nokey,
title = {Closing the Gap between Experiment and Simulation─A Holistic Study on the Complexation of Small Interfering RNAs with Polyethylenimine},
author = {Jonas Binder and Joshua Winkeljann and Katharina Steinegger and Lara Trnovec and Daria Orekhova and Jonas Zähringer and Andreas Hörner and Valentin Fell and Philip Tinnefeld and Benjamin Winkeljann and Wolfgang Frieß and Olivia M Merkel},
url = {https://doi.org/10.1021/acs.molpharmaceut.3c00747},
doi = {10.1021/acs.molpharmaceut.3c00747},
year = {2024},
date = {2024-02-19},
urldate = {2024-02-19},
journal = {Molecular Pharmaceutics},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Fiona Cole; Jonas Zähringer; Johann Bohlen; Tim Schröder; Florian Steiner; Martina Pfeiffer; Patrick Schüler; Fernando D. Stefani; Philip Tinnefeld
Super-resolved FRET and co-tracking in pMINFLUX Journal Article
In: Nature Photonics, 2024.
@article{nokey,
title = {Super-resolved FRET and co-tracking in pMINFLUX},
author = {Fiona Cole and Jonas Zähringer and Johann Bohlen and Tim Schröder and Florian Steiner and Martina Pfeiffer and Patrick Schüler and Fernando D. Stefani and Philip Tinnefeld},
url = {https://doi.org/10.1038/s41566-024-01384-4},
doi = {10.1038/s41566-024-01384-4},
year = {2024},
date = {2024-02-09},
urldate = {2024-02-09},
journal = {Nature Photonics},
keywords = {},
pubstate = {published},
tppubtype = {article}
}

Lennart Grabenhorst; Flurin Sturzenegger; Moa Hasler; Benjamin Schuler; Philip Tinnefeld
Single-Molecule FRET at 10 MHz Count Rates Journal Article
In: J Am Chem Soc, 2024, ISSN: 1520-5126.
@article{pmid38266173,
title = {Single-Molecule FRET at 10 MHz Count Rates},
author = {Lennart Grabenhorst and Flurin Sturzenegger and Moa Hasler and Benjamin Schuler and Philip Tinnefeld},
doi = {10.1021/jacs.3c13757},
issn = {1520-5126},
year = {2024},
date = {2024-01-01},
urldate = {2024-01-01},
journal = {J Am Chem Soc},
abstract = {A bottleneck in many studies utilizing single-molecule Förster resonance energy transfer is the attainable photon count rate, as it determines the temporal resolution of the experiment. As many biologically relevant processes occur on time scales that are hardly accessible with currently achievable photon count rates, there has been considerable effort to find strategies to increase the stability and brightness of fluorescent dyes. Here, we use DNA nanoantennas to drastically increase the achievable photon count rates and observe fast biomolecular dynamics in the small volume between two plasmonic nanoparticles. As a proof of concept, we observe the coupled folding and binding of two intrinsically disordered proteins, which form transient encounter complexes with lifetimes on the order of 100 μs. To test the limits of our approach, we also investigated the hybridization of a short single-stranded DNA to its complementary counterpart, revealing a transition path time of 17 μs at photon count rates of around 10 MHz, which is an order-of-magnitude improvement compared to the state of the art. Concomitantly, the photostability was increased, enabling many seconds long megahertz fluorescence time traces. Due to the modular nature of the DNA origami method, this platform can be adapted to a broad range of biomolecules, providing a promising approach to study previously unobservable ultrafast biophysical processes.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
2023
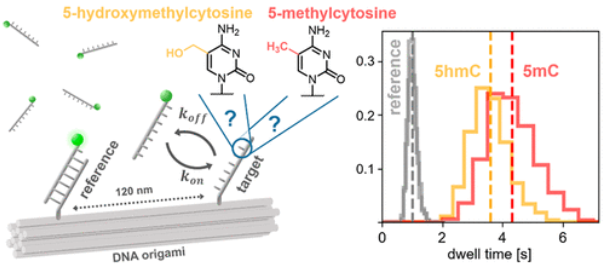
Julian Bauer; Andreas Reichl; Philip Tinnefeld
In: ACS Nano, 2023.
@article{nokey,
title = {Kinetic Referencing Allows Identification of Epigenetic Cytosine Modifications by Single-Molecule Hybridization Kinetics and Superresolution DNA-PAINT Microscopy},
author = {Julian Bauer and Andreas Reichl and Philip Tinnefeld},
doi = {10.1021/acsnano.3c08451},
year = {2023},
date = {2023-12-29},
urldate = {2023-12-29},
journal = {ACS Nano},
abstract = {We develop a DNA origami-based internal kinetic referencing system with a colocalized reference and target molecule to provide increased sensitivity and robustness for transient binding kinetics. To showcase this, we investigate the subtle changes in binding strength of DNA oligonucleotide hybrids induced by cytosine modifications. These cytosine modifications, especially 5-methylcytosine but also its oxidized derivatives, have been increasingly studied in the context of epigenetics. Recently revealed correlations of epigenetic modifications and disease also render them interesting biomarkers for early diagnosis. Internal kinetic referencing allows us to probe and compare the influence of the different epigenetic cytosine modifications on the strengths of 7-nucleotide long DNA hybrids with one or two modified nucleotides by single-molecule imaging of their transient binding, revealing subtle differences in binding times. Interestingly, the influence of epigenetic modifications depends on their position in the DNA strand, and in the case of two modifications, effects are additive. The sensitivity of the assay indicates its potential for the direct detection of epigenetic disease markers.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}

Lars Richter; Alan M Szalai; C Lorena Manzanares-Palenzuela; Izabela Kamińska; Philip Tinnefeld
Exploring the Synergies of Single-Molecule Fluorescence and 2D Materials Coupled by DNA Journal Article
In: Adv Mater, pp. e2303152, 2023, ISSN: 1521-4095.
@article{pmid37670535,
title = {Exploring the Synergies of Single-Molecule Fluorescence and 2D Materials Coupled by DNA},
author = {Lars Richter and Alan M Szalai and C Lorena Manzanares-Palenzuela and Izabela Kamińska and Philip Tinnefeld},
doi = {10.1002/adma.202303152},
issn = {1521-4095},
year = {2023},
date = {2023-09-01},
urldate = {2023-09-01},
journal = {Adv Mater},
pages = {e2303152},
abstract = {The world of 2D materials is steadily growing, with numerous researchers attempting to discover, elucidate, and exploit their properties. Approaches relying on the detection of single fluorescent molecules offer a set of advantages, for instance, high sensitivity and specificity, that allow the drawing of conclusions with unprecedented precision. Herein, it is argued how the study of 2D materials benefits from fluorescence-based single-molecule modalities, and vice versa. A special focus is placed on DNA, serving as a versatile adaptor when anchoring single dye molecules to 2D materials. The existing literature on the fruitful combination of the two fields is reviewed, and an outlook on the additional synergies that can be created between them provided.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
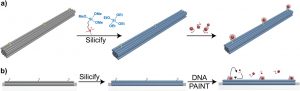
Lea M. Wassermann; Michael Scheckenbach; Anna V. Baptist; Viktorija Glembockyte; Amelie Heuer-Jungemann
Full Site-Specific Addressability in DNA Origami-Templated Silica Nanostructures Journal Article
In: Advanced Materials, 2023.
@article{nokey,
title = {Full Site-Specific Addressability in DNA Origami-Templated Silica Nanostructures},
author = {Lea M. Wassermann and Michael Scheckenbach and Anna V. Baptist and Viktorija Glembockyte and Amelie Heuer-Jungemann},
url = {https://onlinelibrary.wiley.com/doi/10.1002/adma.202212024},
doi = {10.1002/adma.202212024},
year = {2023},
date = {2023-04-25},
urldate = {2023-04-25},
journal = {Advanced Materials},
abstract = {DNA nanotechnology allows for the fabrication of nanometer-sized objects with high precision and selective addressability as a result of the programmable hybridization of complementary DNA strands. Such structures can template the formation of other materials, including metals and complex silica nanostructures, where the silica shell simultaneously acts to protect the DNA from external detrimental factors. However, the formation of silica nanostructures with site-specific addressability has thus far not been explored. Here, it is shown that silica nanostructures templated by DNA origami remain addressable for post silicification modification with guest molecules even if the silica shell measures several nm in thickness. The conjugation of fluorescently labeled oligonucleotides is used to different silicified DNA origami structures carrying a complementary ssDNA handle as well as DNA-PAINT super-resolution imaging to show that ssDNA handles remain unsilicified and thus ensure retained addressability. It is also demonstrated that not only handles, but also ssDNA scaffold segments within a DNA origami nanostructure remain accessible, allowing for the formation of dynamic silica nanostructures. Finally, the power of this approach is demonstrated by forming 3D DNA origami crystals from silicified monomers. These results thus present a fully site-specifically addressable silica nanostructure with complete control over size and shape.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}

Ganesh Agam; Christian Gebhardt; Milana Popara; Rebecca Mächtel; Julian Folz; Benjamin Ambrose; Neharika Chamachi; Sang Yoon Chung; Timothy D Craggs; Marijn de Boer; Dina Grohmann; Taekjip Ha; Andreas Hartmann; Jelle Hendrix; Verena Hirschfeld; Christian G Hübner; Thorsten Hugel; Dominik Kammerer; Hyun-Seo Kang; Achillefs N Kapanidis; Georg Krainer; Kevin Kramm; Edward A Lemke; Eitan Lerner; Emmanuel Margeat; Kirsten Martens; Jens Michaelis; Jaba Mitra; Gabriel G Moya Muñoz; Robert B Quast; Nicole C Robb; Michael Sattler; Michael Schlierf; Jonathan Schneider; Tim Schröder; Anna Sefer; Piau Siong Tan; Johann Thurn; Philip Tinnefeld; John van Noort; Shimon Weiss; Nicolas Wendler; Niels Zijlstra; Anders Barth; Claus A M Seidel; Don C Lamb; Thorben Cordes
Reliability and accuracy of single-molecule FRET studies for characterization of structural dynamics and distances in proteins Journal Article
In: Nat Methods, 2023, ISSN: 1548-7105.
@article{pmid36973549,
title = {Reliability and accuracy of single-molecule FRET studies for characterization of structural dynamics and distances in proteins},
author = {Ganesh Agam and Christian Gebhardt and Milana Popara and Rebecca Mächtel and Julian Folz and Benjamin Ambrose and Neharika Chamachi and Sang Yoon Chung and Timothy D Craggs and Marijn de Boer and Dina Grohmann and Taekjip Ha and Andreas Hartmann and Jelle Hendrix and Verena Hirschfeld and Christian G Hübner and Thorsten Hugel and Dominik Kammerer and Hyun-Seo Kang and Achillefs N Kapanidis and Georg Krainer and Kevin Kramm and Edward A Lemke and Eitan Lerner and Emmanuel Margeat and Kirsten Martens and Jens Michaelis and Jaba Mitra and Gabriel G Moya Muñoz and Robert B Quast and Nicole C Robb and Michael Sattler and Michael Schlierf and Jonathan Schneider and Tim Schröder and Anna Sefer and Piau Siong Tan and Johann Thurn and Philip Tinnefeld and John van Noort and Shimon Weiss and Nicolas Wendler and Niels Zijlstra and Anders Barth and Claus A M Seidel and Don C Lamb and Thorben Cordes},
doi = {10.1038/s41592-023-01807-0},
issn = {1548-7105},
year = {2023},
date = {2023-03-01},
urldate = {2023-03-01},
journal = {Nat Methods},
abstract = {Single-molecule Förster-resonance energy transfer (smFRET) experiments allow the study of biomolecular structure and dynamics in vitro and in vivo. We performed an international blind study involving 19 laboratories to assess the uncertainty of FRET experiments for proteins with respect to the measured FRET efficiency histograms, determination of distances, and the detection and quantification of structural dynamics. Using two protein systems with distinct conformational changes and dynamics, we obtained an uncertainty of the FRET efficiency ≤0.06, corresponding to an interdye distance precision of ≤2 Å and accuracy of ≤5 Å. We further discuss the limits for detecting fluctuations in this distance range and how to identify dye perturbations. Our work demonstrates the ability of smFRET experiments to simultaneously measure distances and avoid the averaging of conformational dynamics for realistic protein systems, highlighting its importance in the expanding toolbox of integrative structural biology.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
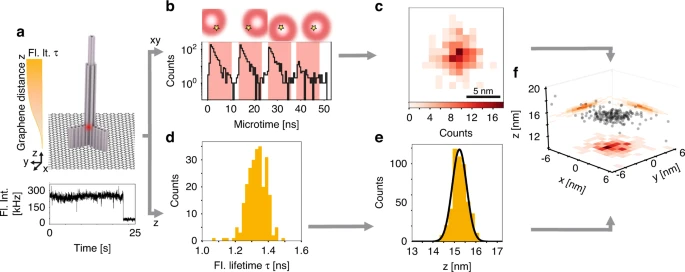
Jonas Zähringer; Fiona Cole; Johann Bohlen; Florian Steiner; Izabela Kamińska; Philip Tinnefeld
Combining pMINFLUX, graphene energy transfer and DNA-PAINT for nanometer precise 3D super-resolution microscopy Journal Article
In: Light Sci Appl, vol. 12, no. 1, pp. 70, 2023, ISSN: 2047-7538.
@article{pmid36898993,
title = {Combining pMINFLUX, graphene energy transfer and DNA-PAINT for nanometer precise 3D super-resolution microscopy},
author = {Jonas Zähringer and Fiona Cole and Johann Bohlen and Florian Steiner and Izabela Kamińska and Philip Tinnefeld},
doi = {10.1038/s41377-023-01111-8},
issn = {2047-7538},
year = {2023},
date = {2023-03-01},
urldate = {2023-03-01},
journal = {Light Sci Appl},
volume = {12},
number = {1},
pages = {70},
abstract = {3D super-resolution microscopy with nanometric resolution is a key to fully complement ultrastructural techniques with fluorescence imaging. Here, we achieve 3D super-resolution by combining the 2D localization of pMINFLUX with the axial information of graphene energy transfer (GET) and the single-molecule switching by DNA-PAINT. We demonstrate <2 nm localization precision in all 3 dimension with axial precision reaching below 0.3 nm. In 3D DNA-PAINT measurements, structural features, i.e., individual docking strands at distances of 3 nm, are directly resolved on DNA origami structures. pMINFLUX and GET represent a particular synergetic combination for super-resolution imaging near the surface such as for cell adhesion and membrane complexes as the information of each photon is used for both 2D and axial localization information. Furthermore, we introduce local PAINT (L-PAINT), in which DNA-PAINT imager strands are equipped with an additional binding sequence for local upconcentration improving signal-to-background ratio and imaging speed of local clusters. L-PAINT is demonstrated by imaging a triangular structure with 6 nm side lengths within seconds.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}

Ece Büber; Tim Schröder; Michael Scheckenbach; Mihir Dass; Henri G Franquelim; Philip Tinnefeld
DNA Origami Curvature Sensors for Nanoparticle and Vesicle Size Determination with Single-Molecule FRET Readout Journal Article
In: ACS Nano, 2023, ISSN: 1936-086X.
@article{pmid36735241,
title = {DNA Origami Curvature Sensors for Nanoparticle and Vesicle Size Determination with Single-Molecule FRET Readout},
author = {Ece Büber and Tim Schröder and Michael Scheckenbach and Mihir Dass and Henri G Franquelim and Philip Tinnefeld},
doi = {10.1021/acsnano.2c11981},
issn = {1936-086X},
year = {2023},
date = {2023-02-01},
urldate = {2023-02-01},
journal = {ACS Nano},
abstract = {Particle size is an important characteristic of materials with a direct effect on their physicochemical features. Besides nanoparticles, particle size and surface curvature are particularly important in the world of lipids and cellular membranes as the cell membrane undergoes conformational changes in many biological processes which leads to diverging local curvature values. On account of that, it is important to develop cost-effective, rapid and sufficiently precise systems that can measure the surface curvature on the nanoscale that can be translated to size for spherical particles. As an alternative approach for particle characterization, we present flexible DNA nanodevices that can adapt to the curvature of the structure they are bound to. The curvature sensors use Fluorescence Resonance Energy Transfer (FRET) as the transduction mechanism on the single-molecule level. The curvature sensors consist of segmented DNA origami structures connected via flexible DNA linkers incorporating a FRET pair. The activity of the sensors was first demonstrated with defined binding to different DNA origami geometries used as templates. Then the DNA origami curvature sensors were applied to measure spherical silica beads having different size, and subsequently on lipid vesicles. With the designed sensors, we could reliably distinguish different sized nanoparticles within a size range of 50-300 nm as well as the bending angle range of 50-180°. This study helps with the development of more advanced modular-curvature sensing devices that are capable of determining the sizes of nanoparticles and biological complexes.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}

Tim Schröder; Johann Bohlen; Sarah E Ochmann; Patrick Schüler; Stefan Krause; Don C Lamb; Philip Tinnefeld
Shrinking gate fluorescence correlation spectroscopy yields equilibrium constants and separates photophysics from structural dynamics Journal Article
In: Proc Natl Acad Sci U S A, vol. 120, no. 4, pp. e2211896120, 2023, ISSN: 1091-6490.
@article{pmid36652471,
title = {Shrinking gate fluorescence correlation spectroscopy yields equilibrium constants and separates photophysics from structural dynamics},
author = {Tim Schröder and Johann Bohlen and Sarah E Ochmann and Patrick Schüler and Stefan Krause and Don C Lamb and Philip Tinnefeld},
doi = {10.1073/pnas.2211896120},
issn = {1091-6490},
year = {2023},
date = {2023-01-01},
urldate = {2023-01-01},
journal = {Proc Natl Acad Sci U S A},
volume = {120},
number = {4},
pages = {e2211896120},
abstract = {Fluorescence correlation spectroscopy is a versatile tool for studying fast conformational changes of biomolecules especially when combined with Förster resonance energy transfer (FRET). Despite the many methods available for identifying structural dynamics in FRET experiments, the determination of the forward and backward transition rate constants and thereby also the equilibrium constant is difficult when two intensity levels are involved. Here, we combine intensity correlation analysis with fluorescence lifetime information by including only a subset of photons in the autocorrelation analysis based on their arrival time with respect to the excitation pulse (microtime). By fitting the correlation amplitude as a function of microtime gate, the transition rate constants from two fluorescence-intensity level systems and the corresponding equilibrium constants are obtained. This shrinking-gate fluorescence correlation spectroscopy (sg-FCS) approach is demonstrated using simulations and with a DNA origami-based model system in experiments on immobilized and freely diffusing molecules. We further show that sg-FCS can distinguish photophysics from dynamic intensity changes even if a dark quencher, in this case graphene, is involved. Finally, we unravel the mechanism of a FRET-based membrane charge sensor indicating the broad potential of the method. With sg-FCS, we present an algorithm that does not require prior knowledge and is therefore easily implemented when an autocorrelation analysis is carried out on time-correlated single-photon data.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}

Kateryna Trofymchuk; Karol Kołątaj; Viktorija Glembockyte; Fangjia Zhu; Guillermo P Acuna; Tim Liedl; Philip Tinnefeld
Gold Nanorod DNA Origami Antennas for 3 Orders of Magnitude Fluorescence Enhancement in NIR Journal Article
In: ACS Nano, 2023, ISSN: 1936-086X.
@article{pmid36594816,
title = {Gold Nanorod DNA Origami Antennas for 3 Orders of Magnitude Fluorescence Enhancement in NIR},
author = {Kateryna Trofymchuk and Karol Kołątaj and Viktorija Glembockyte and Fangjia Zhu and Guillermo P Acuna and Tim Liedl and Philip Tinnefeld},
doi = {10.1021/acsnano.2c09577},
issn = {1936-086X},
year = {2023},
date = {2023-01-01},
urldate = {2023-01-01},
journal = {ACS Nano},
abstract = {DNA origami has taken a leading position in organizing materials at the nanoscale for various applications such as manipulation of light by exploiting plasmonic nanoparticles. We here present the arrangement of gold nanorods in a plasmonic nanoantenna dimer enabling up to 1600-fold fluorescence enhancement of a conventional near-infrared (NIR) dye positioned at the plasmonic hotspot between the nanorods. Transmission electron microscopy, dark-field spectroscopy, and fluorescence analysis together with numerical simulations give us insights on the heterogeneity of the observed enhancement values. The size of our hotspot region is ∼12 nm, granted by using the recently introduced design of NAnoantenna with Cleared HotSpot (NACHOS), which provides enough space for placing of tailored bioassays. Additionally, the possibility to synthesize nanoantennas in solution might allow for production upscaling.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
2022

Kevin N. Baumann, Tim Schröder , Prashanth S. Ciryam, Diana Morzy, Philip Tinnefeld, Tuomas P. J. Knowles, Silvia Hernández-Ainsa
DNA–Liposome Hybrid Carriers for Triggered Cargo Release Journal Article
In: ACS Applied Bio Materials, vol. 5, iss. 8, 2022.
@article{nokey,
title = {DNA–Liposome Hybrid Carriers for Triggered Cargo Release},
author = {Kevin N. Baumann, Tim Schröder
, Prashanth S. Ciryam, Diana Morzy, Philip Tinnefeld,
Tuomas P. J. Knowles, Silvia Hernández-Ainsa},
doi = {doi: 10.1021/acsabm.2c00225},
year = {2022},
date = {2022-07-15},
urldate = {2022-07-15},
journal = {ACS Applied Bio Materials},
volume = {5},
issue = {8},
publisher = {American Chemical Society},
keywords = {},
pubstate = {published},
tppubtype = {article}
}

Cindy Close; Kateryna Trofymchuk; Lennart Grabenhorst; Birka Lalkens; Viktorija Glembockyte; Philip Tinnefeld
Maximizing the Accessibility in DNA Origami Nanoantenna Plasmonic Hotspots Journal Article
In: Advanced Materials Interfaces, vol. 9, iss. 24, pp. 2200255, 2022.
@article{Close2022,
title = {Maximizing the Accessibility in DNA Origami Nanoantenna Plasmonic Hotspots},
author = {Cindy Close and Kateryna Trofymchuk and Lennart Grabenhorst and Birka Lalkens and Viktorija Glembockyte and Philip Tinnefeld},
url = {https://doi.org/10.1002/admi.202200255},
doi = {10.1002/admi.202200255},
year = {2022},
date = {2022-07-01},
urldate = {2022-07-01},
journal = {Advanced Materials Interfaces},
volume = {9},
issue = {24},
pages = {2200255},
publisher = {Wiley},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
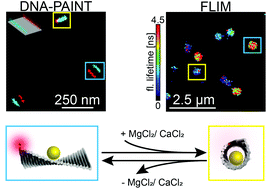
Kristina Hübner; Mario Raab; Johann Bohlen; Julian Bauer; Philip Tinnefeld
Salt-Induced Conformational Switching of a Flat Rectangular DNA Origami Structure Journal Article
In: Nanoscale, vol. 14, pp. 7898 - 7905, 2022.
@article{nokey,
title = {Salt-Induced Conformational Switching of a Flat Rectangular DNA Origami Structure},
author = {Kristina Hübner and Mario Raab and Johann Bohlen and Julian Bauer and Philip Tinnefeld },
url = {https://doi.org/10.1039/D1NR07793G},
doi = {10.1039/D1NR07793G},
year = {2022},
date = {2022-05-11},
urldate = {2022-05-11},
journal = {Nanoscale},
volume = {14},
pages = { 7898 - 7905},
abstract = {A rectangular DNA origami structure is one of the most studied and often used motif for
applications in DNA nanotechnology. Here, we present two assays to study structural
changes in DNA nanostructures and reveal a reversible rolling-up of the rectangular DNA
origami structure induced by bivalent cations such as magnesium or calcium. First, we
applied one-color and two-color superresolution DNA-PAINT with protruding strands
along the long edges of the DNA origami rectangle. At increasing salt concentration, a
single line instead of two lines is observed as a first indicator of rolling-up. Two-color
measurements also revealed different conformations with parallel and angled edges.
Second, we placed a gold nanoparticle and a dye molecule at different positions on the
DNA origami structure. Distance dependent fluorescence quenching by the nanoparticle
reports on dynamic transitions as well as it provides evidence that the rolling-up occurs
preferentially along the diagonal of the DNA origami rectangle. The results will be helpful
to test DNA structural models and the assays presented will be useful to study further
structural transitions in DNA nanotechnology. },
keywords = {},
pubstate = {published},
tppubtype = {article}
}
applications in DNA nanotechnology. Here, we present two assays to study structural
changes in DNA nanostructures and reveal a reversible rolling-up of the rectangular DNA
origami structure induced by bivalent cations such as magnesium or calcium. First, we
applied one-color and two-color superresolution DNA-PAINT with protruding strands
along the long edges of the DNA origami rectangle. At increasing salt concentration, a
single line instead of two lines is observed as a first indicator of rolling-up. Two-color
measurements also revealed different conformations with parallel and angled edges.
Second, we placed a gold nanoparticle and a dye molecule at different positions on the
DNA origami structure. Distance dependent fluorescence quenching by the nanoparticle
reports on dynamic transitions as well as it provides evidence that the rolling-up occurs
preferentially along the diagonal of the DNA origami rectangle. The results will be helpful
to test DNA structural models and the assays presented will be useful to study further
structural transitions in DNA nanotechnology.

Sarah E Ochmann; Tim Schröder; Clara M Schulz; Philip Tinnefeld
Quantitative Single-Molecule Measurements of Membrane Charges with DNA Origami Sensors Journal Article
In: Anal Chem, 2022, ISSN: 1520-6882.
@article{pmid35089694,
title = {Quantitative Single-Molecule Measurements of Membrane Charges with DNA Origami Sensors},
author = {Sarah E Ochmann and Tim Schröder and Clara M Schulz and Philip Tinnefeld},
doi = {10.1021/acs.analchem.1c05092},
issn = {1520-6882},
year = {2022},
date = {2022-01-01},
urldate = {2022-01-01},
journal = {Anal Chem},
abstract = {Charges in lipid head groups generate electrical surface potentials at cell membranes, and changes in their composition are involved in various signaling pathways, such as T-cell activation or apoptosis. Here, we present a DNA origami-based sensor for membrane surface charges with a quantitative fluorescence read-out of single molecules. A DNA origami plate is equipped with modifications for specific membrane targeting, surface immobilization, and an anionic sensing unit consisting of single-stranded DNA and the dye ATTO542. This unit is anchored to a lipid membrane by the dye ATTO647N, and conformational changes of the sensing unit in response to surface charges are read out by fluorescence resonance energy transfer between the two dyes. We test the performance of our sensor with single-molecule fluorescence microscopy by exposing it to differently charged large unilamellar vesicles. We achieve a change in energy transfer of ∼10% points between uncharged and highly charged membranes and demonstrate a quantitative relation between the surface charge and the energy transfer. Further, with autocorrelation analyses of confocal data, we unravel the working principle of our sensor that is switching dynamically between a membrane-bound state and an unbound state on the timescale of 1-10 ms. Our study introduces a complementary sensing system for membrane surface charges to previously published genetically encoded sensors. Additionally, the single-molecule read-out enables investigations of lipid membranes on the nanoscale with a high spatial resolution circumventing ensemble averaging.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
2021

Merve-Zeynep Kesici; Philip Tinnefeld; Andrés Manuel Vera
A simple and general approach to generate photoactivatable DNA processing enzymes Journal Article
In: Nucleic Acids Res, 2021, ISSN: 1362-4962.
@article{pmid34904657,
title = {A simple and general approach to generate photoactivatable DNA processing enzymes},
author = {Merve-Zeynep Kesici and Philip Tinnefeld and Andrés Manuel Vera},
doi = {10.1093/nar/gkab1212},
issn = {1362-4962},
year = {2021},
date = {2021-12-01},
urldate = {2021-12-01},
journal = {Nucleic Acids Res},
abstract = {DNA processing enzymes, such as DNA polymerases and endonucleases, have found many applications in biotechnology, molecular diagnostics, and synthetic biology, among others. The development of enzymes with controllable activity, such as hot-start or light-activatable versions, has boosted their applications and improved the sensitivity and specificity of the existing ones. However, current approaches to produce controllable enzymes are experimentally demanding to develop and case-specific. Here, we introduce a simple and general method to design light-start DNA processing enzymes. In order to prove its versatility, we applied our method to three DNA polymerases commonly used in biotechnology, including the Phi29 (mesophilic), Taq, and Pfu polymerases, and one restriction enzyme. Light-start enzymes showed suppressed polymerase, exonuclease, and endonuclease activity until they were re-activated by an UV pulse. Finally, we applied our enzymes to common molecular biology assays and showed comparable performance to commercial hot-start enzymes.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}

Tim Schröder; Sebastian Bange; Jakob Schedlbauer; Florian Steiner; John M Lupton; Philip Tinnefeld; Jan Vogelsang
How Blinking Affects Photon Correlations in Multichromophoric Nanoparticles Journal Article
In: ACS Nano, 2021, ISSN: 1936-086X.
@article{pmid34735135,
title = {How Blinking Affects Photon Correlations in Multichromophoric Nanoparticles},
author = {Tim Schröder and Sebastian Bange and Jakob Schedlbauer and Florian Steiner and John M Lupton and Philip Tinnefeld and Jan Vogelsang},
doi = {10.1021/acsnano.1c06649},
issn = {1936-086X},
year = {2021},
date = {2021-11-01},
urldate = {2021-11-01},
journal = {ACS Nano},
abstract = {A single chromophore can only emit a maximum of one single photon per excitation cycle. This limitation results in a phenomenon commonly referred to as photon antibunching (pAB). When multiple chromophores contribute to the fluorescence measured, the degree of pAB has been used as a metric to "count" the number of chromophores. But the fact that chromophores can switch randomly between bright and dark states also impacts pAB and can lead to incorrect chromophore numbers being determined from pAB measurements. By both simulations and experiment, we demonstrate how pAB is affected by independent and collective chromophore blinking, enabling us to formulate universal guidelines for correct interpretation of pAB measurements. We use DNA-origami nanostructures to design multichromophoric model systems that exhibit either independent or collective chromophore blinking. Two approaches are presented that can distinguish experimentally between these two blinking mechanisms. The first one utilizes the different excitation intensity dependence on the blinking mechanisms. The second approach exploits the fact that collective blinking implies energy transfer to a quenching moiety, which is a time-dependent process. In pulsed-excitation experiments, the degree of collective blinking can therefore be altered by time gating the fluorescence photon stream, enabling us to extract the energy-transfer rate to a quencher. The ability to distinguish between different blinking mechanisms is valuable in materials science, such as for multichromophoric nanoparticles like conjugated-polymer chains as well as in biophysics, for example, for quantitative analysis of protein assemblies by counting chromophores.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}

Sarah E. Ochmann; Himanshu Joshi; Ece Büber; Henri G. Franquelim; Pierre Stegemann; Barbara Saccà; Ulrich F. Keyser; Aleksei Aksimentiev; Philip Tinnefeld
DNA Origami Voltage Sensors for Transmembrane Potentials with Single-Molecule Sensitivity Journal Article
In: Nano Letters, vol. 21, no. 20, pp. 8634–8641, 2021.
@article{Ochmann2021,
title = {DNA Origami Voltage Sensors for Transmembrane Potentials with Single-Molecule Sensitivity},
author = {Sarah E. Ochmann and Himanshu Joshi and Ece Büber and Henri G. Franquelim and Pierre Stegemann and Barbara Saccà and Ulrich F. Keyser and Aleksei Aksimentiev and Philip Tinnefeld},
doi = {10.1021/acs.nanolett.1c02584},
year = {2021},
date = {2021-10-01},
urldate = {2021-10-01},
journal = {Nano Letters},
volume = {21},
number = {20},
pages = {8634--8641},
publisher = {American Chemical Society (ACS)},
keywords = {},
pubstate = {published},
tppubtype = {article}
}

Martina Pfeiffer; Kateryna Trofymchuk; Simona Ranallo; Francesco Ricci; Florian Steiner; Fiona Cole; Viktorija Glembockyte; Philip Tinnefeld
Single antibody detection in a DNA origami nanoantenna Journal Article
In: iScience, vol. 24, no. 9, pp. 103072, 2021.
@article{Pfeiffer2021,
title = {Single antibody detection in a DNA origami nanoantenna},
author = {Martina Pfeiffer and Kateryna Trofymchuk and Simona Ranallo and Francesco Ricci and Florian Steiner and Fiona Cole and Viktorija Glembockyte and Philip Tinnefeld},
doi = {10.1016/j.isci.2021.103072},
year = {2021},
date = {2021-09-01},
urldate = {2021-09-01},
journal = {iScience},
volume = {24},
number = {9},
pages = {103072},
publisher = {Elsevier BV},
keywords = {},
pubstate = {published},
tppubtype = {article}
}

Viktorija Glembockyte; Lennart Grabenhorst; Kateryna Trofymchuk; Philip Tinnefeld
DNA Origami Nanoantennas for Fluorescence Enhancement Journal Article
In: Acc Chem Res, vol. 54, no. 17, pp. 3338–3348, 2021.
@article{Glembockyte2021,
title = {DNA Origami Nanoantennas for Fluorescence Enhancement},
author = {Viktorija Glembockyte and Lennart Grabenhorst and Kateryna Trofymchuk and Philip Tinnefeld},
doi = {10.1021/acs.accounts.1c00307},
year = {2021},
date = {2021-08-01},
urldate = {2021-08-01},
journal = {Acc Chem Res},
volume = {54},
number = {17},
pages = {3338--3348},
publisher = {American Chemical Society (ACS)},
keywords = {},
pubstate = {published},
tppubtype = {article}
}

Andrés Manuel Vera; Albert Galera-Prat; Michał Wojciechowski; Bartosz Różycki; Douglas V. Laurents; Mariano Carrión-Vázquez; Marek Cieplak; Philip Tinnefeld
Cohesin-dockerin code in cellulosomal dual binding modes and its allosteric regulation by proline isomerization Journal Article
In: Structure, vol. 29, no. 6, pp. 587–597.e8, 2021.
@article{Vera2021,
title = {Cohesin-dockerin code in cellulosomal dual binding modes and its allosteric regulation by proline isomerization},
author = {Andrés Manuel Vera and Albert Galera-Prat and Michał Wojciechowski and Bartosz Różycki and Douglas V. Laurents and Mariano Carrión-Vázquez and Marek Cieplak and Philip Tinnefeld},
doi = {10.1016/j.str.2021.01.006},
year = {2021},
date = {2021-06-01},
urldate = {2021-06-01},
journal = {Structure},
volume = {29},
number = {6},
pages = {587--597.e8},
publisher = {Elsevier BV},
keywords = {},
pubstate = {published},
tppubtype = {article}
}

Izabela Kamińska; Johann Bohlen; Renukka Yaadav; Patrick Schüler; Mario Raab; Tim Schröder; Jonas Zähringer; Karolina Zielonka; Stefan Krause; Philip Tinnefeld
Graphene Energy Transfer for Single-Molecule Biophysics, Biosensing, and Super-Resolution Microscopy Journal Article
In: Advanced Materials, vol. 33, no. 24, pp. 2101099, 2021.
@article{Kaminska2021,
title = {Graphene Energy Transfer for Single-Molecule Biophysics, Biosensing, and Super-Resolution Microscopy},
author = {Izabela Kamińska and Johann Bohlen and Renukka Yaadav and Patrick Schüler and Mario Raab and Tim Schröder and Jonas Zähringer and Karolina Zielonka and Stefan Krause and Philip Tinnefeld},
doi = {10.1002/adma.202101099},
year = {2021},
date = {2021-05-01},
urldate = {2021-05-01},
journal = {Advanced Materials},
volume = {33},
number = {24},
pages = {2101099},
publisher = {Wiley},
keywords = {},
pubstate = {published},
tppubtype = {article}
}

Stefan Krause; Evelyn Ploetz; Johann Bohlen; Patrick Schüler; Renukka Yaadav; Florian Selbach; Florian Steiner; Izabela Kamińska; Philip Tinnefeld
Graphene-on-Glass Preparation and Cleaning Methods Characterized by Single-Molecule DNA Origami Fluorescent Probes and Raman Spectroscopy Journal Article
In: ACS Nano, vol. 15, no. 4, pp. 6430–6438, 2021.
@article{Krause2021,
title = {Graphene-on-Glass Preparation and Cleaning Methods Characterized by Single-Molecule DNA Origami Fluorescent Probes and Raman Spectroscopy},
author = {Stefan Krause and Evelyn Ploetz and Johann Bohlen and Patrick Schüler and Renukka Yaadav and Florian Selbach and Florian Steiner and Izabela Kamińska and Philip Tinnefeld},
doi = {10.1021/acsnano.0c08383},
year = {2021},
date = {2021-04-01},
urldate = {2021-04-01},
journal = {ACS Nano},
volume = {15},
number = {4},
pages = {6430--6438},
publisher = {American Chemical Society (ACS)},
keywords = {},
pubstate = {published},
tppubtype = {article}
}

Kristina Hübner; Himanshu Joshi; Aleksei Aksimentiev; Fernando D. Stefani; Philip Tinnefeld; Guillermo P. Acuna
Determining the In-Plane Orientation and Binding Mode of Single Fluorescent Dyes in DNA Origami Structures Journal Article
In: ACS Nano, vol. 15, no. 3, pp. 5109–5117, 2021.
@article{Huebner2021,
title = {Determining the In-Plane Orientation and Binding Mode of Single Fluorescent Dyes in DNA Origami Structures},
author = {Kristina Hübner and Himanshu Joshi and Aleksei Aksimentiev and Fernando D. Stefani and Philip Tinnefeld and Guillermo P. Acuna},
doi = {10.1021/acsnano.0c10259},
year = {2021},
date = {2021-03-01},
urldate = {2021-03-01},
journal = {ACS Nano},
volume = {15},
number = {3},
pages = {5109--5117},
publisher = {American Chemical Society (ACS)},
keywords = {},
pubstate = {published},
tppubtype = {article}
}

Alan M. Szalai; Bruno Siarry; Jerónimo Lukin; Sebastián Giusti; Nicolás Unsain; Alfredo Cáceres; Florian Steiner; Philip Tinnefeld; Damián Refojo; Thomas M. Jovin; Fernando D. Stefani
Super-resolution Imaging of Energy Transfer by Intensity-Based STED-FRET Journal Article
In: Nano Letters, vol. 21, no. 5, pp. 2296–2303, 2021.
@article{Szalai2021,
title = {Super-resolution Imaging of Energy Transfer by Intensity-Based STED-FRET},
author = {Alan M. Szalai and Bruno Siarry and Jerónimo Lukin and Sebastián Giusti and Nicolás Unsain and Alfredo Cáceres and Florian Steiner and Philip Tinnefeld and Damián Refojo and Thomas M. Jovin and Fernando D. Stefani},
doi = {10.1021/acs.nanolett.1c00158},
year = {2021},
date = {2021-02-01},
urldate = {2021-02-01},
journal = {Nano Letters},
volume = {21},
number = {5},
pages = {2296--2303},
publisher = {American Chemical Society (ACS)},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
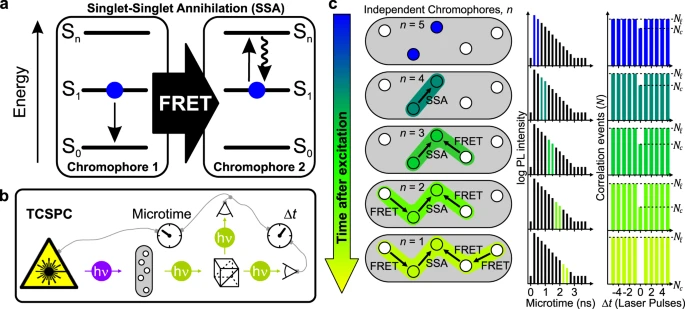
Gordon J. Hedley; Tim Schröder; Florian Steiner; Theresa Eder; Felix J. Hofmann; Sebastian Bange; Dirk Laux; Sigurd Höger; Philip Tinnefeld; John M. Lupton; Jan Vogelsang
Picosecond time-resolved photon antibunching measures nanoscale exciton motion and the true number of chromophores Journal Article
In: Nature Communications, vol. 12, no. 1, 2021.
@article{Hedley2021,
title = {Picosecond time-resolved photon antibunching measures nanoscale exciton motion and the true number of chromophores},
author = {Gordon J. Hedley and Tim Schröder and Florian Steiner and Theresa Eder and Felix J. Hofmann and Sebastian Bange and Dirk Laux and Sigurd Höger and Philip Tinnefeld and John M. Lupton and Jan Vogelsang},
url = {https://www.cup.uni-muenchen.de/de/aktuelles/archiv/2021/energieuebertragung-in-organischen-leuchtdioden-oled/
https://www.e-conversion.de/the-secret-of-the-missing-excitons/},
doi = {10.1038/s41467-021-21474-z},
year = {2021},
date = {2021-02-01},
urldate = {2021-02-01},
journal = {Nature Communications},
volume = {12},
number = {1},
publisher = {Springer Science and Business Media LLC},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
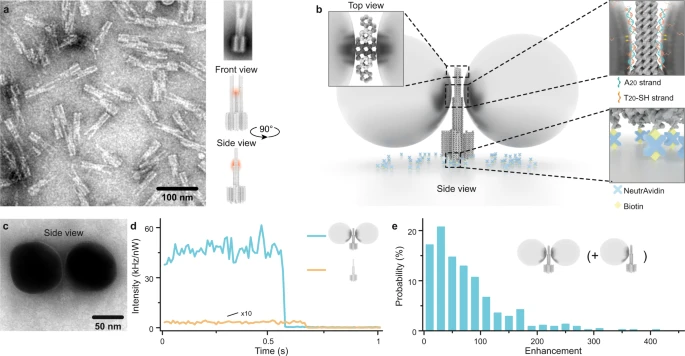
Kateryna Trofymchuk; Viktorija Glembockyte; Lennart Grabenhorst; Florian Steiner; Carolin Vietz; Cindy Close; Martina Pfeiffer; Lars Richter; Max L. Schütte; Florian Selbach; Renukka Yaadav; Jonas Zähringer; Qingshan Wei; Aydogan Ozcan; Birka Lalkens; Guillermo P. Acuna; Philip Tinnefeld
Addressable nanoantennas with cleared hotspots for single-molecule detection on a portable smartphone microscope Journal Article
In: Nature Communications, vol. 12, no. 1, 2021.
@article{Trofymchuk2021,
title = {Addressable nanoantennas with cleared hotspots for single-molecule detection on a portable smartphone microscope},
author = {Kateryna Trofymchuk and Viktorija Glembockyte and Lennart Grabenhorst and Florian Steiner and Carolin Vietz and Cindy Close and Martina Pfeiffer and Lars Richter and Max L. Schütte and Florian Selbach and Renukka Yaadav and Jonas Zähringer and Qingshan Wei and Aydogan Ozcan and Birka Lalkens and Guillermo P. Acuna and Philip Tinnefeld},
doi = {10.1038/s41467-021-21238-9},
year = {2021},
date = {2021-02-01},
urldate = {2021-02-01},
journal = {Nature Communications},
volume = {12},
number = {1},
publisher = {Springer Science and Business Media LLC},
keywords = {},
pubstate = {published},
tppubtype = {article}
}

Patrick Eiring; Ryan McLaughlin; Siddharth S Matikonda; Zhongying Han; Lennart Grabenhorst; Dominic A Helmerich; Mara Meub; Gerti Beliu; Michael Luciano; Venu Bandi; Niels Zijlstra; Zhen-Dan Shi; Sergey G Tarasov; Rolf Swenson; Philip Tinnefeld; Viktorija Glembockyte; Thorben Cordes; Markus Sauer; Martin J Schnermann
Targetable Conformationally Restricted Cyanines Enable Photon-Count-Limited Applications* Journal Article
In: Angew Chem Int Ed Engl, vol. 60, no. 51, pp. 26685–26693, 2021, ISSN: 1521-3773.
@article{pmid34606673,
title = {Targetable Conformationally Restricted Cyanines Enable Photon-Count-Limited Applications*},
author = {Patrick Eiring and Ryan McLaughlin and Siddharth S Matikonda and Zhongying Han and Lennart Grabenhorst and Dominic A Helmerich and Mara Meub and Gerti Beliu and Michael Luciano and Venu Bandi and Niels Zijlstra and Zhen-Dan Shi and Sergey G Tarasov and Rolf Swenson and Philip Tinnefeld and Viktorija Glembockyte and Thorben Cordes and Markus Sauer and Martin J Schnermann},
doi = {10.1002/anie.202109749},
issn = {1521-3773},
year = {2021},
date = {2021-01-01},
urldate = {2021-01-01},
journal = {Angew Chem Int Ed Engl},
volume = {60},
number = {51},
pages = {26685--26693},
abstract = {Cyanine dyes are exceptionally useful probes for a range of fluorescence-based applications, but their photon output can be limited by trans-to-cis photoisomerization. We recently demonstrated that appending a ring system to the pentamethine cyanine ring system improves the quantum yield and extends the fluorescence lifetime. Here, we report an optimized synthesis of persulfonated variants that enable efficient labeling of nucleic acids and proteins. We demonstrate that a bifunctional sulfonated tertiary amide significantly improves the optical properties of the resulting bioconjugates. These new conformationally restricted cyanines are compared to the parent cyanine derivatives in a range of contexts. These include their use in the plasmonic hotspot of a DNA-nanoantenna, in single-molecule Förster-resonance energy transfer (FRET) applications, far-red fluorescence-lifetime imaging microscopy (FLIM), and single-molecule localization microscopy (SMLM). These efforts define contexts in which eliminating cyanine isomerization provides meaningful benefits to imaging performance.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}

Michael Scheckenbach; Tom Schubert; Carsten Forthmann; Viktorija Glembockyte; Philip Tinnefeld
Self-Regeneration and Self-Healing in DNA Origami Nanostructures Journal Article
In: Angewandte Chemie International Edition, vol. 60, no. 9, pp. 4931–4938, 2021.
@article{Scheckenbach2021,
title = {Self-Regeneration and Self-Healing in DNA Origami Nanostructures},
author = {Michael Scheckenbach and Tom Schubert and Carsten Forthmann and Viktorija Glembockyte and Philip Tinnefeld},
doi = {10.1002/anie.202012986},
year = {2021},
date = {2021-01-01},
urldate = {2021-01-01},
journal = {Angewandte Chemie International Edition},
volume = {60},
number = {9},
pages = {4931--4938},
publisher = {Wiley},
keywords = {},
pubstate = {published},
tppubtype = {article}
}